undergraduate authors underlined
2025 – present
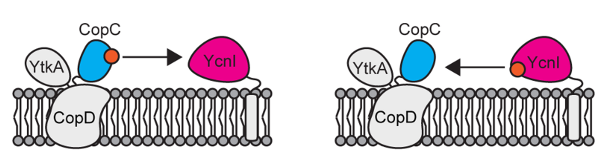
- de Oliveira Silva, Y.R., Barnes, G., Zheng, D., Zhitnitsky, D., Geathers, S.J., Peters, S.C., Szalai, V.A., Helmann, J.D., Fisher, O.S.‡ Copper acquisition in Bacillus subtilis involves Cu(II) exchange between YcnI and YcnJ. 2025. J Biol Chem. doi: https://doi.org/10.1016/j.jbc.2025.110480.
- Calicdan, X., Fisher, O.S., Ha, B.H., Boggon, T.J., Stiegler, A.L. Nickel binding to a split ATCUN motif in c-Src SH3 domain facilitates crystallization. 2025. Prot. Pept. Lett. doi: 10.2174/0109298665417324250929120040.
2020 – 2024
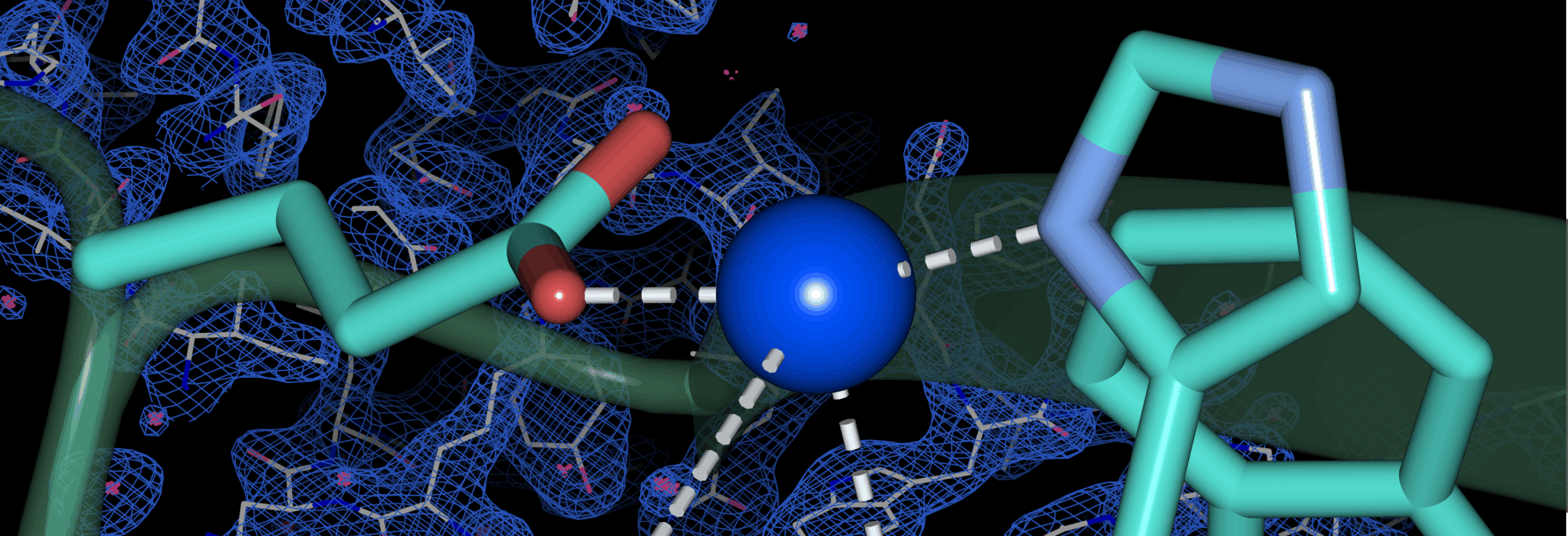
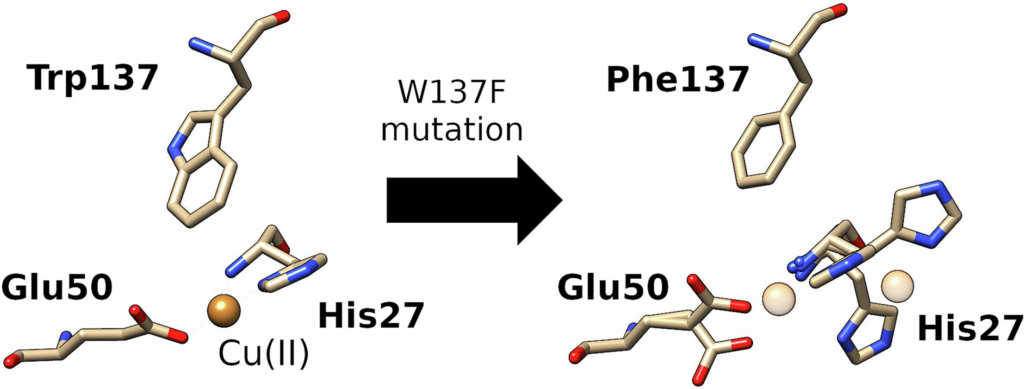
- de Oliveira Silva, Y.R.*, Zheng, D.*, Peters, S.C., Fisher, O.S. Stabilization of a Cu-binding site by a highly conserved tryptophan residue. 2024. J Inorg Biochem. doi: 10.1016/j.jinorgbio.2024.112501.
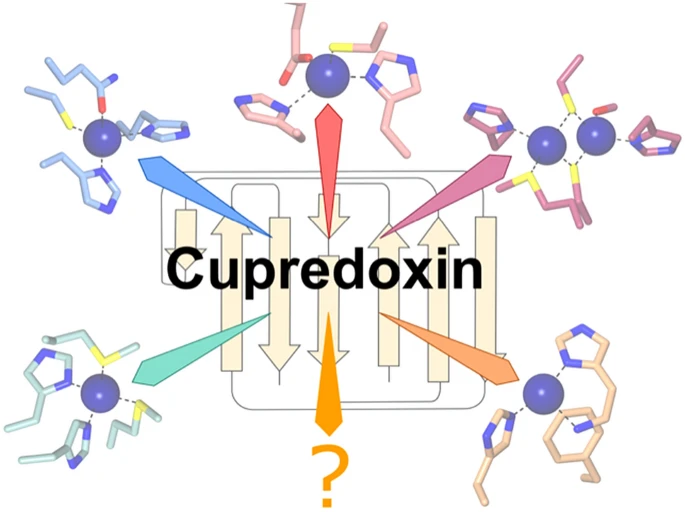
- Guo, J. and Fisher, O.S. Orchestrating copper binding: structure and variations on the cupredoxin fold. 2022. J Biol Inorg Chem. doi: 10.1007/s00775-022-01955-2.
- Damle, M.S., Singh, A.N., Peters, S.C., Szalai, V.A., Fisher, O.S. The YcnI protein from Bacillis subtilis contains a copper-binding domain. 2021. J Biol Chem. Sep;297(3):101078. doi: 10.1016/j.jbc.2021.101078.
- Fisher, O.S.*, Li, X.*, Liu, W.*, Zhang, R.*, Boggon, T.J. Crystallographic studies of the cerebral cavernous malformations proteins. 2020. Methods Mol Biol. 2152, 291-302. doi: 10.1007/978-1-0716-0640-7_21
2016 – 2019
- Fisher, O.S., Sendzik, M.R., Ross, M.O., Lawton, T.J., Hoffman, B.M., Rosenzweig, A.C. PCuAC domains from methane-oxidizing bacteria use a histidine brace to bind copper. 2019. J Biol Chem. Nov 1;294(44):16351-16363. doi: 10.1074/jbc.RA119.010093.
- Miller, C.J., Lou, H.J., Simpson, C., van de Kooij, B., Ha, B.H., Fisher, O.S., Pirman, N.L., Boggon, T.J., Rinehart, J., Yaffe, M.B., Linding, R., Turk, B.E. Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output. 2019. PLoS Biol. 17, e2006540. DOI: 10.1371/journal.pbio.2006540
- Ross, M.O.*, Fisher, O.S.*, Morgada, M.N., Krzyaniak, M.D., Wasielewski, M., Vila, A.J., Hoffman, B.M., Rosenzweig, A.C. Formation and electronic structure of an atypical CuA site. 2019. J Am Chem Soc. 141, 4678-4686. DOI: 10.1021/jacs.8b13610
- Fisher, O.S., Kenney, G.E., Ross, M.O., Ro, S.Y., Lemma, B.E., Batelu, S., Thomas, P.M., Sosnowski, V.C., DeHart, C.J., Kelleher, N.L., Stemmler, T.L., Hoffman, B.M., Rosenzweig, A.C. Characterization of a long overlooked copper protein from methane- and ammonia-oxidizing bacteria. 2018. Nat Commun. 9, 4276. DOI: 10.1038/s41467-018-06681-5
- Wang, W., Nguyen, L.T.T., Burlak, C., Chegini, F., Guo, F., Chataway, T., Ju, S., Fisher, O.S., Miller, D.W., Datta, D., Wu, F., Wu, C., Landeru, A., Wells, J.A., Cookson, M.R., Boxer, M.B., Thomas, C.J., Gai, W., Ringe, D., Petsko, G.A., Hoang, Q.Q. Caspase-1 causes truncation and aggregation of the Parkinson disease-associated protein α-synuclein. 2016. Proc Natl Acad Sci U S A. 113(34), 9587-92. DOI: 10.1073/pnas.1610099113
2015 and earlier
- Li, X., Fisher, O.S., Boggon, T.J. The cerebral cavernous malformations proteins. 2015. Oncotarget. 6, 32279-32280. DOI: 10.18632/oncotarget.5443
- Fisher, O. S.*, Deng, H.*, Liu, D.*, Zhang, Y., Wei, R., Deng, Y. Zhang, F., Louvi, A., Turk, B.E., Boggon, T.J.*, Su, B.*. Structure and vascular function of MEKK3-cerebral cavernous malformations complex. 2015. Nat Commun. 6, 7937. DOI: 10.1038/ncomms8937
- Draheim, K.M.*, Li, X.*, Zhang, R., Fisher, O.S., Villari, G., Calderwood, D.A., Boggon, T.J. Structural determinants of a CCM3:CCM2 interaction that stabilizes protein expression and permits endothelial network formation. 2015. J Cell Biol. 208, 987-1001. DOI: 10.1083/jcb.201407129
- Fisher, O.S., Liu, W., Zhang, R., Stiegler, A.L., Ghedia, S., Weber, J.L., Boggon, T.J. Structural basis for the disruption of the Cerebral Cavernous Malformations 2 (CCM2) interaction with Krev Interaction Trapped 1 (KRIT1) by disease-associated mutations. 2015. J Biol Chem. 290, 2842-53. DOI: 10.1074/jbc.M114.616433
- Fisher, O.S. and Boggon, T.J. Signaling pathways and the cerebral cavernous malformations proteins: lessons from structural biology. 2014. Cell Mol Life Sci 71, 1881-1892. DOI: 10.1242/jcs.138388
- Draheim, K.M., Fisher, O.S., Boggon, T.J., and Calderwood, D.A. Cerebral cavernous malformation proteins at a glance. 2014. J Cell Sci 127, 701-707. DOI: 10.1007/s00018-013-1532-9
- Fisher, O.S., Zhang, R., Li, X., Murphy, J.W., Demeler, B., and Boggon, T.J. Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus. 2013. FEBS Lett 587, 272-277. DOI: 10.1016/j.febslet.2012.12.011