We use a variety of complementary approaches spanning X-ray crystallography, bioinorganic chemistry, biochemistry, biophysics, and microbiology to explore questions related to how bacterial respond and adapt to environmental changes.
Copper homeostasis in Gram positive bacteria
Copper (Cu) is one of the most ancient antimicrobial tools, but there are still major gaps in the fundamental understanding of how bacteria handle this transition metal.
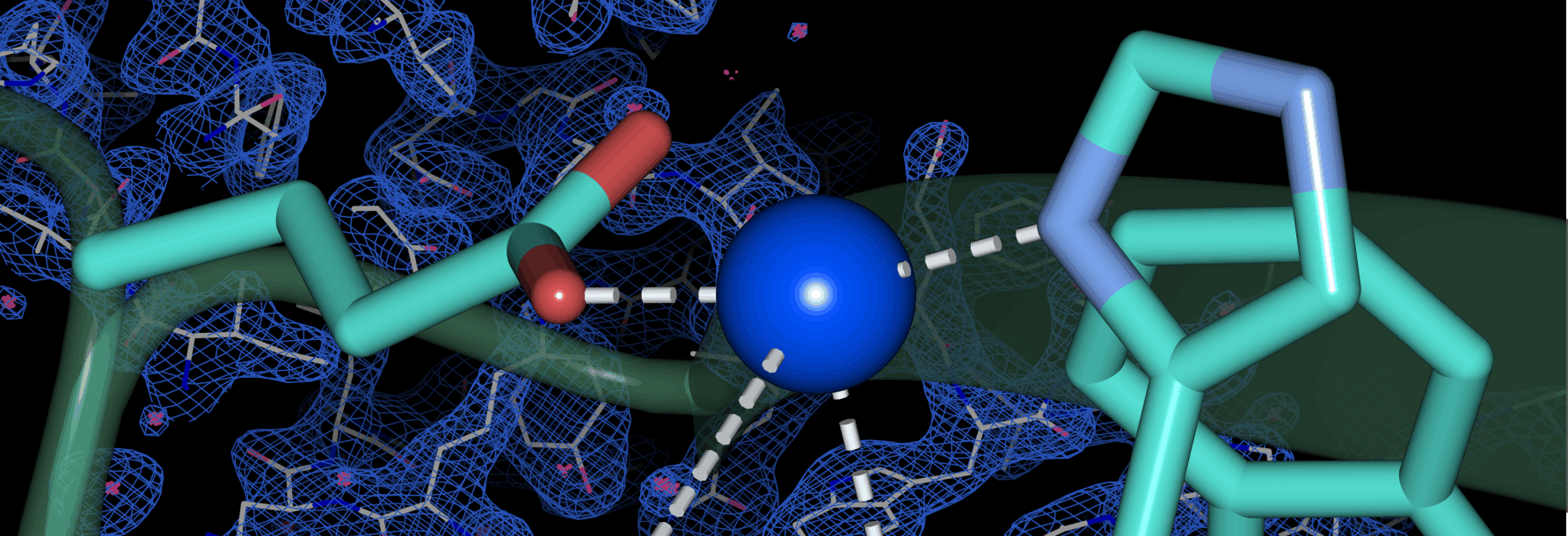
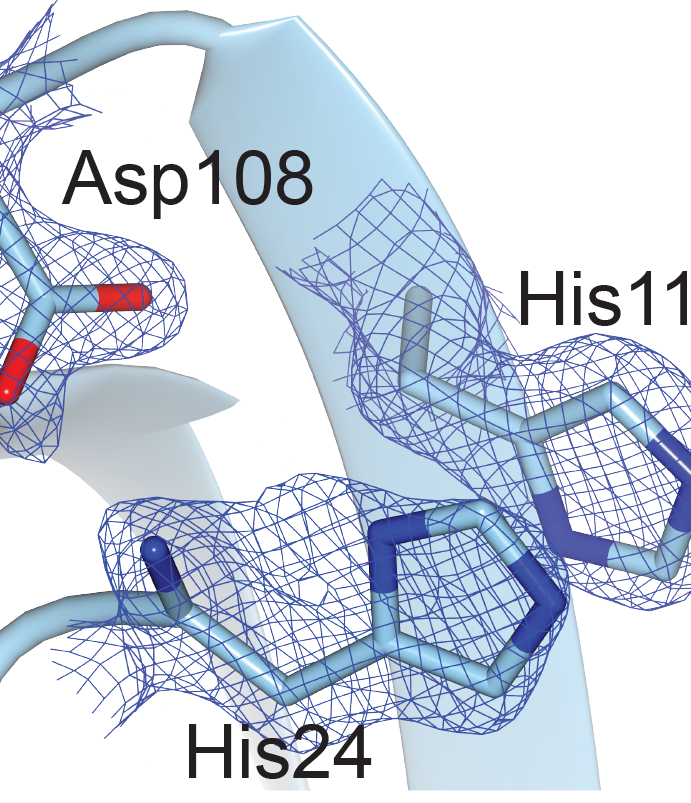
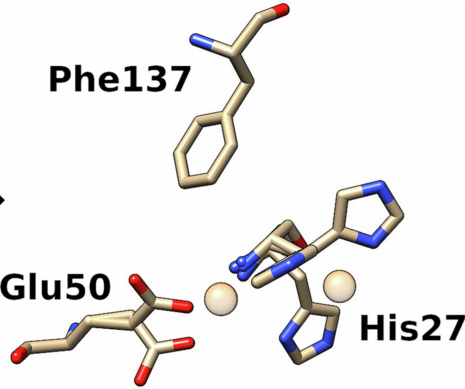
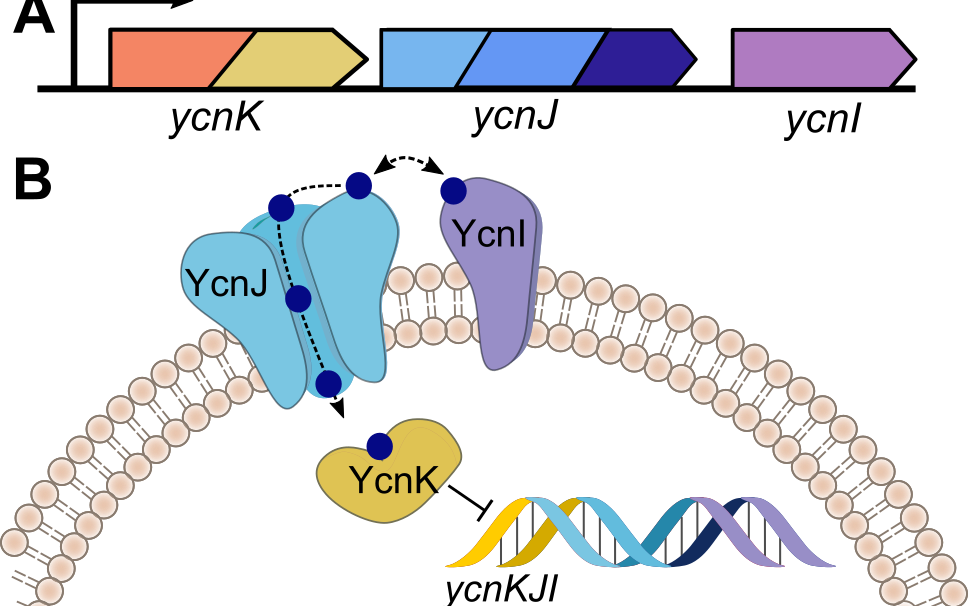
Significant research efforts have greatly advanced the current knowledge of how microorganisms remove Cu when it is in excess. More recently, Cu- dependent proteins have also been found to reside in the cytosol, suggesting that intracellular Cu may be an important component of bacterial life. We hypothesize that Cu uptake is regulated by Cu-dependent transcriptional repressors and the proteins under their control. To investigate this question, we are currently focusing on understanding the structure, biochemical properties, and physiological roles of the proteins encoded by the ycn operon from Bacillus subtilis and their interactions with copper ions.
Recent publications:
Drivers of bacterial enzyme specificity
In many signal transduction pathways, kinases transfer phosphate groups to specific substrates, yet for some subclasses of bacterial kinases, how this specificity is conferred remains poorly understood.
Bacteria can mount responses to different types of stimuli using one or more kinases that phosphorylate their substrates. A subclass of these enzymes are found across bacterial phylogeny, yet carry out distinct chemistry from most of their bacterial counterparts. What features of these enzymes drives their specificity for their preferred substrates? We hypothesize that structural variations are important for conferring distinct substrate specificity and kinetics and can be targeted by small molecule inhibitors. To investigate this question, we are currently focusing on using bioinformatics, biochemistry, and structural biology to understand the evolutionary history of a subset of serine kinases and how they recognize and interact with their substrates.
Recent publications:
Coming soon!